Evolving

This website works best at 67% zoom.

Welcome to our lab page!
Image credits: Arkive, O. Sanisidro, Mark Garlick/Science Photo Library, Davide Bonadonna/National Geographic, Christian Jegou/Science Photo Library
The vast majority of Life that existed on our planet is extinct, leaving behind only a handful of surviving lineages with a remarkably conservative molecular makeup: proteins, lipids, and sugars generate all organismal form and function and encode information on physiology and relationships (phylogeny).
Modern biology relies primarily on molecular -omics data to decipher organismal relationships, identify developmental drivers of evolution, and elucidate how organisms interact with each other and their environment. However, integrable biomolecular information has traditionally been assumed to be lost during fossilization, leaving vast gaps in our understanding of the long-term evolutionary drivers shaping the diversity, physiology, and ecology of contemporary life forms. Consequently, our ability to forecast how modern biodiversity will respond to our changing planet in the long run is critically limited by mechanistic inferences from the most recent snapshot of Life’s evolutionary history.
Our lab is based in the Morton K. Blaustein Department of Earth & Planetary Sciences at Johns Hopkins University. We combine expertise from chemistry, biology, and geology to bridge the ‘Molecular Gap’ between Life past and present, ultimately aiming to elucidate the foundational molecular mechanisms of evolution currently concealed within the geological record. We have pioneered the field of Molecular Paleobiology by demonstrating the mechanistic linkage between biomacromolecules and insoluble kerogen preserved in deepest time, and identified the first time-inclusive biomolecular signatures: my team employs experimental, empirical, and theoretical approaches applied to modern and fossil biological samples, curated in museums and collected in the field, to track the alterations of informative biomacromolecules through time and space. By leveraging these insights, we develop molecular phylogenetic and physiological biosignatures that can be integrated across both modern and extinct life forms – generating new primary data that effectively allow us to fill the blank pages in the history of Life.
Our main objective is to investigate evolutionary responses to global environmental and ecological turnovers: using an integrative approach, we target the most critical intervals in Life’s evolutionary history to gather insights that otherwise could not be inferred from the biased sample of lineages surviving into the modern. We not only endeavor to develop solutions for carbon capture inspired by the geological principles of organic matter alteration and storage, but also translate the principles of physiological evolution into models that predict the long-term effects of climate change on today's biodiversity. Hopkins offers an incredibly welcoming and intellectually stimulating environment with complementary expertise and analytical facilities present in Biology, Physics & Astronomy, Chemistry, Engineering, and the Medical School.
Our work spans from prebiotic chemistry to the diversity of Life as we (don't ?) know it, through time and space – we always welcome creative and ambitious ideas, new challenges, and complementary methodological expertise! If this sounds exciting to you, make sure to check out opportunities in our lab and reach out!









Research


1 Biomolecule fossilization
Our previous research revealed that proteins, lipids, sugars, and certain pigments leave characteristic traces in the fossil record. Different classes of biomolecules undergo characteristic chemical alterations during diagenesis, and tend to transform into more stable compounds which can survive in the geological record.
2 Crosslinking as the mechanism of preservation
During early diagenesis (days to years postmortem), oxidative conditions trigger the formation of Reactive Carbonyl Species (RCS) from lipids and sugars. Oxidative conditions are prevalent in, i.e., fluvial, alluvial, and shallow marine sediments. Lipid- and sugar-RCS show distinct affinities for exposed amino acid residues of omnipresent proteins. Primary targets are nucleophilic amino acid residues, such as arginine, lysine, histidine, and cystein, as well as the protein N-terminus. Observed fossilization reactions include primarily C-C-bond forming reactions, such as transition metal-catalyzed additions. The resulting crosslinking products fall either in the category of Advanced Lipoxidation End Products (ALEs), or Advanced Glycoxidation End Products (AGEs). ALEs and AGEs constitute pre-graphitic insoluble kerogen composed of N-, O-, S-heterocyclic polymers – compounds that are water-insoluble, indigestible to microbes, and energetically stable. There does not seem to be an age limit for the preservation of N-, O-, S-heterocyclic polymers in the geological record!
3 Mineral interactions
The geological product of crosslinking reactions, termed insoluble kerogen, often coordinates transition metals that stabilize the resulting coordination complexes against temperature, pressure, and dissolution stress in the rock record. Such organo-mineral interactions are tissue-specific and can facilitate early diagenetic mineralization of biological materials. Mineralization aids in preserving fragile organic morphologies and storing climate-inactive organic carbon in deep time. We have analyzed more than 300 organic-rich fossils and sediments and are currently expanding this sample to understand the mechanisms involved in phosphatization, pyritization, silicification, and clay mineral precipitation on organic templates. We anticipate that these insights will explain observed biases in the fossil record.
Bridging the ‘Molecular Gap’: biomolecule and biosignature fossilization
We explore to which extent we can truly integrate biological signals encoded in the composition of biomolecules across the physical parameter space. Our lab tackles this challenge by leveraging a combination of experimental, empirical, and theoretical approaches to track the alterations of informative biomacromolecules and the kinetics of biosignature loss as a function of variables (pressure, temperature, etc.) that are relevant to fossilization processes on Earth and elsewhere in the solar system. Our team critically interrogates residual molecular phylogenetic and physiological signals revealed through comparative geochemistry with complementary genomic inferences. We are passionate methodologists, and enjoy developing new protocols, hardware, and data analysis tools for end users interested in the application of our integrative proxies.

New data, new opportunities:
the first time-integrative molecular biosignatures and their transformative applications
We apply our different integrative biosignatures to the most challenging open questions in the history of Life. With countless opportunities for transformative applications of the molecular biomineralization, tissue identity, metabolic, and phylogenetic signals, we focus our energy on collecting the first molecular mechanistic insights into evolutionary transitions of primary societal and economic relevance. We find inspiration in our discoveries of mechanisms facilitating molecular preservation in deep time, and adaptations of extinct organisms to carbon fixation during past hot-house climates, and translate these insights into carbon capture and storage technology. We enjoy venturing into uncharted scientific territories and help defining the frontier questions at the interface of the fundamental sciences. Research in our lab is very dynamic and catalyzed by the expertise and ideas of our team members!
Below, you can find a brief introduction of the four biomacromolecular signatures that currently guide our scientific endeavors. In addition, there is a snapshot of current research priorities in the lab related to big questions in astrobiology, prebiotic chemistry, organismal evolution and ecology, biodiversity conservation, and new solutions to carbon capture.
We are incredibly grateful to have brilliant collaborators working with us on our various projects - at Hopkins, in Maryland, and across North America, Australia, South America, Europe, Africa, and Asia!

1 Biomineralization signal
The most prominent signal present in compositional fingerprints of biological materials and their fossilization products identifies templated biomineralization.
In a templated biomineral, coordinating amino acid residues interact with ions of the mineral phase. Non-biomineralized tissues lack mineral-templating proteins and comparable, abundant organo-mineral interactions. This signal preserves accurately even in temperature- and pressure-matured carbonaceous compression fossils from the Precambrian and Cambrian (the time window capturing the diversification of animal skeletons), and accurately distinguishes diagenetically and biologically precipitated minerals.
This proxy has the potential to fundamentally advance our understanding of the nature and evolution of biomineralized tissues, as exemplified in our recent study on eggshell evolution in Nature.

2 Tissue identity signal
The tissue type signal groups tissues by identity based on their specific relative abundancies of proteins, lipids, and sugars. Using a training data set of tissues with known identity, modern and fossil tissues of unknown type can efficiently and non-destructively be characterized.
Different tissues contain characteristic relative amounts of proteins, lipids, and sugars - these compositional fingerprints preserve in the relative amounts of lipoxidation and glycoxidation end products: during early diagenesis, lipids generate characteristic Reactive Carbonyl Species (RCS) which differ from those formed by sugars. Depending on their relative abundance in a tissue, lipid- and sugar-derived RCS crosslink with omnipresent proteins, preserving the original abundance of proteins, lipids, and sugars in the resulting composition of N-, O-, S-heterocyclic polymers in a fossil. This is the second most prominent signal in an animal tissue ChemoSpace.
Being able to unambiguously identify tissues in modern and fossil samples can provide informative characters that help resolving phylogenetic affinities, as shown in our recent study on the phylogenetic affinity of the enigmatic tully monster, published in Geobiology.

3 Metabolic rate signal
Metabolic stress leaves molecular traces that preserve in deep time and can be quantified to reconstruct the metabolic rates of both extant and extinct animals. Vascularized and cellular tissues, such as bone, preserve metabolic modifications in their proteinaceous phase.
The metabolic proxy requires the use of homologous structures (due to intraskeletal, tissue-dependent variation). Ongoing research focuses on expanding the phylogenetic applicability of the proxy and discriminating between minute differences in metabolic signals (i.e., true endotherms versus seasonal endotherms). Aerobic mitochondrial respiration is far older than the amniote clade, and we thus explore the manifestation of molecular metabolic signals in invertebrates and extinct prokayotes.
Once calibrated based on skeletal metabolic markers of living animals, the abundance of metabolic stress markers can be translated into a metabolic rate (= the amount of oxygen respired relative to the body mass, in one hour of time).

4 Phylogenetic signal
The phylogenetic signal relies on the compositional differences of amino acid-specific crosslinks in N-, O-, S-heterocyclic polymers. Even though no sequence information can be obtained, the similarity in the relative amino acid composition of homologous tissues provides insights into animal relationships.
Proteins represent translation products of the genetic code. A codon composed of a nucleotide triplet encodes for one out of 21 (for eukaryotes, 22 for all organisms) proteinogenic amino acids. Sequencing modern proteins yields information on organismal relationships, however, diagenetic alteration of peptide bonds during fossilization prevents the sequential cleavage of units through any conventional method. Amine- and thiol-bearing amino acid residues yield diagnostic fossilization products. When comparing N-, O-, S-heterocyclic polymers of the same tissue type and from similar depositional settings, a cluster analysis of the fossil composition reflects organismal relationships. The ability to complement morphological assessments of organismal relationships with molecular data has the potential to resolve controversial nodes in the tree of life.
Space, the final frontier: molecular ecosystems across the solar system and a world before the first complex cells
Past: How did Life emerge, interact, and evolve on early Earth?
What were the selective drivers impacting the composition of pre-cellular ecosystems?
Present & Future: Can we define a fingerprint of Life as we know it to improve our search for Life beyond Earth?
Can we constrain plausible chemistries of Life as we don't know it?
Endless forms most beautiful: the many ways Life makes mineral skeletons
Present: What is the relationship between organic template and precipitated biomineral?
Past: Can we use the biomineralization signal to elucidate when and why the first skeletons evolved?
How did different biomineralizers react to changing ocean geochemistry (as function of climatic excursions) in the past?
Future: Can extinct biomineralizers from past hot-house climates inspire carbon capture technology?



We use extraterrestrial insoluble organic matter imported by carbonaceous meteorites (from Mars, the asteroid belt, and other solar system bodies) as an abiogenic comparison for modern and fossil biogenic organic matter. Statistical analyses of compositional data allow us to define the molecular fingerprint shared by all Life as we know it, and reveal compositional features (apomorphies) that are diagnostic of increasing complexity (ranging from prebiotic mixtures to the first cellular structures preserved in the fossil record on Earth).
We combine techniques allowing for molecular characterization with in situ observed experimental pressure- and temperature-alteration and theoretical models based on physical variables, to elucidate the origin(s), diversity, and evolution of earliest Life on Earth and elsewhere in the solar system.
Our group explores the foundational mechanisms of templated biomineralization across modern prokaryote and eukaryote systems to reveal how different protein features translate into mineralogies. For modern biominerals, we integrate data on the proteomic make-up, molecular structures, and spatially resolved post-translational modifications in the context of organo-mineral interactions. We experimentally model the pressure- and temperature-alteration (relevant for fossilization processes) of biominerals, and refine our proxies to allow for the integration of data for modern and fossil samples.
We leverage these insights to unravel [1] the evolutionary mechanisms involved in the independent emergence of biominerals and [2] their clade-specific adaptation potential to long-term climatic and ecological perturbations, with special emphasis on marine calcifiers involved in carbon fixation. Our group translates this information into bio-inspired climate solutions.


Life finds a way: do physiological and reproductive trade-offs determine the fate of organisms during mass extinctions?
Present: Can we refine the metabolic proxy and apply it taxon-inclusively from single-celled protists to cellular vertebrate bone?
Past: How does the energetic trade-off between metabolism and reproduction relate to major environmental perturbations?
How do metabolic changes throughout development/ontogeny impact survival of mass extinction events?
Future: Can these mechanistic insights improve models predicting the long-term impact of climate change on modern biodiversity?


Organisms focus their energetic investments either on their metabolism (to grow faster and larger) or reproduction (increased hatchling survival due to parental care). It is the trade-off between metabolism and reproduction that shapes ecological niches and determines extinction susceptibility during times of global perturbations. Until recently, we were unable to gather molecular metabolic and reproductive behavioral data from extinct organisms.
Combining genomic, spatial molecular, and developmental data for living groups with integrable metabolic and reproductive signals from fossils, bridged through experimental fossilization, we work to reveal the evolutionary relationship and selective drivers (environmental, ecological, ontogenetic) of metabolism and reproduction in a comparative framework, across clades. We apply mechanistic insights to more accurately predict the long-term impact of climate change on modern biodiversity.
P/T mass extinction
K/Pg mass extinction
Research in our lab is proudly supported by:

Interdisciplinary
Consortia for
Astrobiology
Research [ICAR]

Publications
post - HOP: July 2024 and onwards
Stay tuned for exciting research from the PaLEO Lab!
23. Coming soon!
22. Wong, M. L., Prabhu, A., Alexander, C. O’D., Cleaves II, H. J., Cody, G. D., Hystad, G., Bermanec, M., Bleeker, W., Boyce, C. K., Corpolongo, A., Czaja, A., Das, S., Gaines, R. R., Gregory, D., Jaszczak, J., Javaux, E. J., Jodder, J., Knoll, A. H., Van Kranendonk, M., Maloney, K. M., Noffke, N., Rainbird, R., Slaughter, E., Stüeken, E. E., Summons, R. E., Westall, F., Wiemann, J., Xiao, S., & Hazen, R. M. Organic geochemical evidence for life in Archean rocks identified by pyrolysis-GC-MS and supervised machine learning. PNAS, 2025. Click here!


While our lab spaces are being renovated, my team and I have developed a variety of machine-learning and AI tools for the extraction, denoising, and prediction of biosignatures ... stay tuned!

21. Tripp, M.*, Wiemann, J., Brosnan, L., I., Rickard, W. D. A., Vajda, V., Bottcher, M. E., Greenwood, P. F., & Grice, K. Mineralization controls informative biomarker preservation associated with soft part preservation in deep time. Geobiology 23, e70030 (2025). Click here!
20. Wiemann, J.* Fossil dinosaur cells – 20 years after a discovery. Nature (2025). Click here!
PNAS
pre - HOP: 2015 to July 2024
Publications prior to starting my lab at Johns Hopkins
19. Wiemann, J.*, and Heck, P. R. Quantifying the impact of sample, instrument, and data processing on biological signatures detected with Raman spectroscopy. Journal of Raman Spectroscopy, early view. Click here!
18. Wiemann, J.*, Menendez, I.*, Crawford, J.M., Fabbri, M., Gauthier, J.A., Hull, P.M., Norell, M.A., and Briggs, D.E.G. RE: Comment on amniote metabolism and the evolution of endothermy. Nature 621, E4–E6 (2023). Click here!
17. Tripp, M.*, Wiemann, J., Brocks, J., Mayer, P., Lidgard, S., Schwark, L., and Grice, K. Molecular evidence from coprolites reveals dietary strategies of the Carboniferous Mazon Creek fauna. Biology.
– Invited contribution (special issue). Click here!
16. Norell, M. A., Wiemann, J.*, Menendez, I.*, Fabbri, M.*, Yu, C., Marsicano, C., Pol, D., Moore-Nall, A., Varricchio, D. J., and Zelenitsky, D. Cross-methodological evidence confirms soft eggs in sauropodomorph dinosaurs. Nature. Click here!
15. Wiemann, J.*, Menendez, I., Crawford, J.M., Fabbri, M., Gauthier, J.A., Hull, P.M., Norell, M.A., and Briggs, D.E.G. Fossil biomolecules reveal an avian metabolism in the ancestral dinosaur. Nature, 10.1038/s41586-022-04770-6. Altmetric score: 2033. [Dissertation chapter 3/5] – Most widely shared paleontology paper in 2022. Click here!
14. Wiemann, J.* and Briggs, D.E.G. Raman spectroscopy is a powerful tool in a range of paleobiological applications. Invited contribution, Bioessays, 10.1002/bies.202100070. Altmetric score: 11. Click here!
Preprint: Wiemann, J.* and Briggs, D.E.G. Validation of biosignatures confirms the informative nature of fossil organic Raman spectra. BioRxiv. Click here!
13. Wiemann, J.*, Crawford, J.M., and Briggs, D.E.G., 2020. Phylogenetic and physiological signals in metazoan fossil biomolecules. Science Advances, 10.1126/sciadv.aba6883. Altmetric score: 103. Click here!
12. Norell, M. A.*, Wiemann, J.*, Fabbri, M.*, Yu, C., Marsicano, C., Pol, D., Moore-Nall, A., Varricchio, D. J., and Zelenitsky, D., 2020. The first dinosaur egg was soft. Nature. Altmetric score: 1439. Click here!
11. McCoy, V. E.*, Wiemann, J.*, Lamsdell, J.C., Whalen, C.D., Lidgard, S., Mayer, P., Petermann, H., and Briggs, D.E.G., 2020. Chemical signatures of soft tissues distinguish between vertebrates and invertebrates from the Carboniferous Mazon Creek Lagerstätte of Illinois. Geobiology, 10.1111/gbi.12397. Altmetric score: 179. Click here!
10. Ibrahim, N., Maganuco, S., Dal Sasso, C., Fabbri, M., Auditore, M., Bindellini, G., Martill, D.M., Wiemann, J., Zouhri, S., Matarelli, D., Unwin, D.M., Joger, U., Amane, A., Jakubczak, J., Bonadonna, D., Lauder, G., and Pierce S., 2020. Tail-propelled aquatic locomotion in a theropod dinosaur. Nature, 10.1038/s41586-020-2190-3. Altmetric score: 3780. Click here!
9. Wiemann, J.*, de Queiroz, K., Rowe, T. B., Planavsky, N. J., Anderson, R. P., Gogarten, J. P., Turner, P. E., and Gauthier, J. A., 2020. Biota. In: de Queiroz, K., Cantino, P. D., and Gauthier, J. A., Editors. Phylonyms: a companion to the PhyloCode. Berkeley, University of California Press - Invited contribution. Registration number: 298. Click here!
8. Wiemann, J.*, de Queiroz, K., Rowe, T. B., Planavsky, N. J., Anderson, R. P., Gogarten, J. P., Turner, P. E., and Gauthier, J. A., 2020. Pan-Biota. In: de Queiroz, K., Cantino, P. D., and Gauthier, J. A., Editors. Phylonyms: a companion to the PhyloCode. Berkeley, University of California Press - Invited contribution. Registration number: 299. Click here!
7. Fabbri, M., Wiemann, J., Manucci, F., Briggs, D.E.G., 2020. 3-D soft tissue preservation revealed in the skin of a non-avian dinosaur. Palaeontology, 10.1111/pala.12470. Altmetric score: 53. Click here!
6. Yang, T.-R., Wiemann, J., Xu, L., Cheng, Y.-N., Wu, X.-C., and Sander, P. M., 2019. Reconstruction of oviraptorid clutches illuminates their unique nesting biology. Acta Palaeontologica Polonica, 10.4202/app.00597.2018. Click here!
5. Wiemann, J.*, Yang, T.R., and Norell, M.A., 2019. Reply to: Egg pigmentation probably has an archosaurian origin. Nature, 10.1038/s41586-019-1283-3. Altmetric score: 35. Click here!
4. Wiemann, J.*, Fabbri, M., Yang, T.R., Stein, K., Sander, P.M., Norell, M.A., and Briggs, D.E.G., 2018. Fossilization transforms vertebrate hard tissue proteins into N-heterocyclic polymers. Nature Communications, 10.1038/s41467-018-07013-3. Altmetric score: 200. Click here!
3. Wiemann, J.*, Yang, T.R., and Norell, M.A., 2018. Dinosaur egg color had a single evolutionary origin. Nature, 10.1038/s41586- 018-0646-5. Altmetric score: 1624. Click here!
2. Yang, T.R., Chen, Y.-H., Wiemann, J., Spiering, B., and Sander, P.M., 2018. Fossil eggshell cuticle elucidates dinosaur nesting ecology. PeerJ, 6, p. e5144. Click here!
1. Wiemann, J.*, Yang, T.R., Sander, P.N., Schneider, M., Engeser, M., Kath-Schorr, S., Müller, C.E. and Sander, P.M., 2017. Dinosaur origin of egg color: oviraptors laid blue-green eggs. PeerJ, 5, p.e3706. Altmetric score: 461. Click here!























Facilities






















Facilities at Hopkins: FIB, SEM, TEM (I + II), EDX, EBSD, microCT, Bio- & Tissue Printer, Sequencing, etc.
PaLEO labs (EPS): [1] Spectroscopy lab (UV/Vis/NIR AFM NanoRaman + TERS lab) and [2] chemical - BSL 2 wet lab
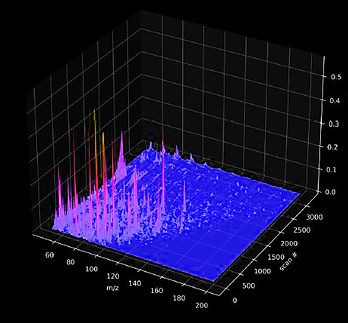
We are currently operating our customized Raman microspectrometer in a temporary lab space based in Bloomberg Physics & Astronomy. Our permanent laser lab and the chemical and microbiological (BSL 2) wet lab and 3-D printing hub are currently being renovated - both spaces are located in the Morton K. Blaustein Department of Earth and Planetary Sciences. Raman spectroscopy allows the non-destructive, holistic chemical characterization of complex materials by probing diagnostic interactions between light and molecular or crystal lattice vibrations. Raman spectroscopy is very time-efficient and facilitates the explorative analysis of samples with unknown composition, such as insoluble organic matter preserved in fossils. Large and integrative data sets can be built in a relatively short amount of time for both biological, geological, paleontological, and meteoritic samples. The resulting spectra are particularly suitable for statistical analyses aiding in the search for biosignatures.
After 10 years of exploring different spectrometer types and specifications, I have chosen a Horiba Jobin-Yvon base model which has been tailored to meet our specific analytical needs as cross-disciplinary scientists who work with inherently complex biocomposite materials: our instrument has three different lasers installed, covering the ultraviolet, visible green, and near-infrared portions of the electromagnetic spectrum! All three lasers can be used in environmental microscopy mode (for 2D, 3D, and 4D mapping), and our visible lasers can additionally be operated in the AFM-mode (= atomic force microscopy; for molecular resolution in spatial compound maps). Spatially and temporally resolved compound maps can be collected (label-free!) by taking advantage of the confocal capabilities of our instrument. A hydrothermal diamond anvil cell can be fitted into the microscope framework and allows us to perform experimental pressure-, temperature-, and fluid-based alteration of samples, observable in situ. To capture potentially informative heterogeneities in near-graphitic (high-pressure/high-temperature matured) biomolecule fossilization products, we use SERS (= surface-enhanced Raman spectroscopy) and TERS (= tip-enhanced Raman spectroscopy) for single-molecule characterization employing combined Plasmonic and Raman effects. The various analytical capabilities in our spectroscopy lab represent the current State-of-the-Art!
Curious? Visitors (bringing interesting samples) are always welcome - our Raman system is fully operational, and we anticipate to move into our permanent laboratory spaces in early 2026! Until then, we share our temporary lab space with our wonderful colleagues in Physics & Astronomy.
Labs in EPS: Geochemistry wet lab, rock prepation/histology lab, microscopy lab, various mass spectrometry labs ...
Go ahead and explore the details for these various fancy EPS facilities operated by my lab and my awesome colleagues on our departmental homepage!
-
Our 3-D Printing Hub: resin printer + Prusa Core 1 [PLA] printer
-
JHU zoological and paleontological research collections
Labs at Hopkins: Biology, Physics & Astronomy, Chemistry, MCP (Materials characterization), Medical School (FAE, Sequencing, etc.)
Museum collections: Smithsonian National Museum of Natural History (Washington DC)
American Museum of Natural History (New York)
Follow the links to explore the online catalogues for the NMNH and AMNH collections!
Live animals & conservation in Baltimore: National Aquarium (aquatic metazoans, herpetofauna, tropical mammals and birds)
The Maryland Zoo in Baltimore (diverse metazoans)
Follow the links to explore the homepage of the National Aquarium and the Baltimore Zoo!
The team [We are currently recruiting and evolving - Interested in joining us? Check out opportunities in the lab!]
We are an international group and would be happy to chat with you in various languages - all our team members are fluent in English, but we also speak German, French, Portuguese, Flemish, Spanish, Norwegian, and Russian!
Principal Investigator: Dr. Jasmina Wiemann
(Phonetic spelling guide: YahS-Mee-Naa WhEE-Mun)
I am a Molecular Geo-, Astro-, & Paleobiologist with broad interests in revealing the past, present, and predictable future of Life on Earth and beyond through the exploration and integrative application of molecular biosignatures. I currently work as an Assistant Professor of Earth and Planetary Sciences at Johns Hopkins University and the Johns Hopkins Medical Institute.
My academic journey started at the age of 15, when I was awarded an Excellence Stipend to study Chemistry at the Technical University of Dortmund in Germany. I went on to obtain degrees in the Geosciences (BSc; Honors Program Scholarship) and Organismal and Evolutionary Biology (MSc; Honors Program Scholarship) at the University of Bonn (Germany). During my MPhil and PhD at Yale University in the United States, I combined these disciplines to actively develop the field of Molecular Paleobiology by mechanistically linking informative biomacromolecules to insoluble kerogen preserved in the geological record. My dissertation yielded more than 19 publications (incl. 9x Nature, Science Adv, Nature Comms) demonstrating the preservation of integrative molecular biosignatures in deep time. After graduating, I held Trimble & Barr Postdoctoral Fellowships at Caltech, allowing me to focus on the preservation kinetics and macroevolutionary applications of biological information in terrestrial and extraterrestrial insoluble kerogen. Supported by an Agouron Institute Fellowship hosted at the University of Chicago and the Field Museum, I worked to constrain the molecular fingerprint of Life as we know it, based on computational, experimental, and analytical work. I am a passionate teacher, seasoned mentor (16 undergraduate-to-postdoctoral mentees), and a proud advocate for women in STEM and first-generational academics.

Teaching at Johns Hopkins University
I have taught more than 23 classes/courses/seminars/lectures/workshops on both the undergraduate and graduate student level prior to starting my position at JHU - in my classes, I cover traditional and frontier research concepts and methodologies across the sciences, connected through timely and interesting narratives. I believe that the most useful classes teach transferable methods and critical thinking: foundational skills that enable independent research!
Given that we work at the interface of geology, chemistry, and biology, and employ statistical methods for data analysis, I offer classes that cover all aspects of our work:
Deciphering the history of Life [AS.271.406 (01)] offered in the Morton K. Blaustein of Earth and Planetary Science at Johns Hopkins University.
Instructor: J. Wiemann. MW | 3 hours/week. Chemistry, Organismal & Evolutionary Biology, Statistics.
The Dynamic Earth: An Introduction to Geology [AS.270.220 (01)] offered in the Morton K. Blaustein of Earth and Planetary Science at Johns Hopkins University.
Instructors: D. Viete, J. Wiemann. TTh | 3 hours/week. Geology, Mineralogy.
The Dynamic Earth Laboratory [AS.270.221 (01)] offered in the Morton K. Blaustein of Earth and Planetary Science at Johns Hopkins University.
Instructors: D. Viete, J. Wiemann. W | 2 hours/week. Geology, Mineralogy.
Visiting Faculty & PIs [Interested in joining us for your sabbatical at Hopkins? Feel free to reach out!]
Dr. Tom Andre Nordheim
PI at the JHU Applied Physics Laboratory & Sabbatical Visitor in the PaLEO Lab
Contact info: tom.nordheim@jhuapl.edu
Cross-disciplinary expertise: Planetary Science & Space Missions
Ocean Worlds
Robotics & Remote Sensing
Biosignature detection
Sabbatical project: Understanding the fingerprints and fates of organic carbon
across planetary systems.
.jpg)

Postdoctoral Scholars
Dr. Mariana Valéria Haase
Postdoctoral Scholar in the PaLEO Lab - joining in September 2025
Previous research: Marie Skłodowska-Curie Actions & Emerging Talent Fellow
at Sorbonne Université, Paris, France
PhD, MSc, BSc: Earth Sciences - Federal Rural University of Pernambuco, Brazil
Cross-disciplinary expertise: Paleobiology & Evolution on a Changing Planet
Vertebrate Metabolism & Development
Comparative Histology & Anatomy
Research interests: The impact of ontogenetic trade-offs between metabolism, growth, and reproduction on past and present amniote extinction susceptibility.


Dr. Pjotr Meyvisch
Postdoctoral Scholar in the PaLEO Lab - joining in November 2025
PhD research: Doctoral & Teaching Fellow at Universiteit Gent, Ghent, Belgium
MSc, BSc: Geology - Universiteit Gent, Ghent, Belgium
Cross-disciplinary expertise: Single-cell Vibrational Spectroscopy & Omics
Time-integrative Biosignatures & Proxy Development
Micro-organisms & Primary Producers through Time
Oceanography & Marine Microbial Ecology
Research interests: Integrating single-cell spectroscopy and structural elucidation to develop time-integrative biosignatures that illuminate the evolution of multicellularity.


PhD Researchers

Liam Olden, MRes
2nd - year PhD student from: Perth, Western Australia
Cross-disciplinary expertise: Geology & Paleontology
Sedimentology, Stratigraphy
Geochemistry, Fossilization
Micro- & Astrobiology
Global Change Biology
Research interests: Evolution of biomineralization in prokaryote & eukaryote systems applied to predict adaptability ranges on a changing planet



Megan Miller, BSc
2nd - year PhD student from: Wisconsin, United States of America
Cross-disciplinary expertise: Molecular Biology & Chemistry
Genomics & Development
Microbiology, Evolution
Geology & Paleontology
Global Change Biology
Research interests: Environmental, ecological, & developmental drivers of
physiological evolution across the reptile clade (incl. birds)


2025
Steven Cohen Award
External Graduate Student Research Affiliates
Tom van der Linden, MSc
1st - year PhD student based at: Freie Universitaet Berlin, Germany
Cross-disciplinary expertise: Organic & Isotope Geochemistry
Quantitative Paleobiology
Reproductive Biology
Geology & Taphonomy
Global Change Biology
Collaborates on: Evolution of amniote reproductive innovations and their impact
on survival bias during extinction events in the past and present


Undergraduate Researchers
Emily Pituch
Based at: Johns Hopkins University
Major 1: Earth and Planetary Sciences
Major 2: Molecular, Cellular, and Developmental Biology
Research interests: Characterizing extraterrestrial organic matter from meteorites
(Asteroid belt, Mars, Moon) to delineate a biogenicity signal
Topics: Astrobiology, Prebiotic chemistry, Biosignatures

.jpg)
Ana Sofia Cancio Trelfa
Based at: Johns Hopkins University
Major: Earth and Planetary Sciences
Research interests: Surveying the abundance and nature of molecular, cellular, and soft-tissue preservation in the fossil record
(PaLEO Lab Project - successfully finished: Summer 2024)
Topics: Vertebrate paleontology, Invertebrate paleontology, evolution


Fanny Borukhova
Based at: Johns Hopkins University
Major: Molecular, Cellular, and Developmental Biology
Soon: MS in Public Health at Johns Hopkins School of Public Health
Research interests: Biomarkers through time - illuminating the survival of a
phylogenetic signal in biomolecule fossilization products
Individual research project successfully finished
- manuscript in prep.
Topics: Biomarkers & diagnostics, Evolutionary medicine, Clinical applications


Lab Turtellini

Maxwell and Galilea
External Affiliates
Species: Trachemys scripta (subspecies: elegans) - Red-eared slider turtle
Family: Emydidae
Hatch date: 2018
Interests: Sunbasking (= behavioral thermoregulation in ectotherms)
Topics: Archosauromorpha

Opportunities in the lab - Join us!
SICB
Portland, USA
January 03-07, 2026
LPSC
The Woodlands, USA
March 16-20, 2026
MA - NE Geobiology
Edelman Fossil Park, USA
TBD, 2026
AbSciCon
Wisconsin, USA
May 17-22, 2026
IPC 7
Cape Town, South Africa
Nov. 30 - Dec. 03, 2026
MA Geobiology
Baltimore, USA
February 28, 2025
BD 2025
Athens, Greece
September 24-26, 2025
Origins Federation
Chicago, USA
September 09-12, 2025
GSA
San Antonio, USA
October 19-22, 2025
SVP
Birmingham, UK
November 12-15, 2025
AbSciCon
Providence, USA
May 5-10, 2024
NAPC
Ann Arbor, USA
June 17-21, 2024
IGC
Busan, Korea
August 25-31, 2024
SVP
Minneapolis, USA
October 30-Nov. 02, 2024
SICB
Atlanta, USA
January 03-07, 2025
I am currently recruiting undergraduate students interested in (paid) research opportunities (based at JHU), graduate students, and postdoctoral scholars for my evolving lab! We are a very energetic and universally curious lab united through our shared passion for cross-disciplinary approaches to bold organismal, ecological, and evolutionary questions! If this sounds exciting to you, please reach out to me (jasmina.wiemann@jhu.edu) - for prospective graduate students (click here for details) and postdocs: please add a brief summary of your research interests and, if available, a short CV.
There is a number of amazing fellowships available that allow postdoctoral scholars to join our team, including three types of Prize Fellowships in the Morton K. Blaustein Department of Earth an Planetary Sciences, and various national and international fellowship awards. I would be happy to advise you on funding opportunities and help designing your tailored project!
The most relevant external funding opportunities for research in our field include (but are not limited to) the NSF EAR Postdoctoral Fellowship (for US citizens), the NSF Bio Postdoctoral Fellowship (for US citizens - I am already committed to an applicant for the 2025/2026 cycle), the Schmidt Science Fellowships (requires nomination by your PhD-granting institution), the Fulbright Fellowship program (for international applicants), the MSCA International Postdoctoral Fellowship (in collaboration with a European university), the Human Frontier Science Program Long-Term and Cross-Disciplinary Fellowships (for international applicants), the NASA Astrobiology Postdoctoral Fellowship Program (I hope to be eligible for sponsorship later this year), the Simons Foundation Fellowship, postdoctoral funding through a regular NSF Project Proposal (for international applicants), etc.
If you would like to chat in person or learn more about research in the lab, there might be opportunities to catch up at upcoming meetings: I will be attending a couple of scientific conferences focused on different topics; dates and venues are listed below - I look forward to meeting you!
News from the lab
2026
01
03
2026 is off to a great start: members of the lab are en route to present at SICB in Portland, OR!
01
12
Upcoming AFM-Raman training session for the lab ...
01
20
The teaching term is about to start with a new practical course co-organized by all members of the lab!

2026
2025

02
14
Thanks to our brilliant colleagues for including us in a potentially transformative NASA grant! Submitted!
03
14
Jas made the Top 40 list of Influencers in Paleontology - accessibly communicating our research to the world is so important!
05
01
We got our AFM-Raman platform delivered to our temporary lab space in Physics & Astronomy!
05
25
Congrats to Megan for winning the Steven Cohen Award from the Society of Vertebrate Paleontology for her research!
08
02
Megan and Jas got their talks accepted at SVP and will be visiting the UK in November!
09
12
Pjotr, Megan, and Jas are in Chicago at the Origins Federation meeting: biosignatures yield new evolutionary insights!
10
20
Jas has received a Fellowship from the David & Lucile Packard Foundation - Wohoo!!!
02
28
Congrats to Megan and Liam for presenting outstanding talks on their first thesis chapters at the MA Geobio Meeting!
03
17
First opinion piece for the new lab is published in Nature! 2 pages on why we care about the fate of biomolecules through time!
05
12
Liam got admitted to the International Geobiology Course! Huge congrats and safe travels to Italy!
05
27
Congrats to Jas, Sarah, Alexa, & Alyssa for winning a miniWISE grant to organize the genotype-to-phenotype symposium!
08
15
The AFM-Raman system is fully operational in Bloomberg P&A! Onwards to new discoveries!
09
20
We are officially funded by NASA! Machine learning might improve biosignature detection on planet Earth and beyond!
11
25
Megan, Jas, and Tom presented at SVP in Birmingham (UK); Megan is off to the Isle of Skye!
03
10
Jas received one of the two Packard Nominations at Johns Hopkins University - Wohoo!
03
20
We're getting close to the shipping date for our AFM-Raman system! So excited (spectroscopy-pun intended)!
05
17
Congrats to Megan and Liam for winning JHU EPS field work and museum work grants!
06
01
Congrats to Jas for winning a JHU Teaching grant to set-up the 3D Print Hub in EPS and print our first osteological collection!
09
05
Jas & Liam got their talks accepted at GSA and will present new data on biomineralization!
10
10
Pjotr and Mari both received their visas and will arrive soon in Baltimore! Samples are already waiting to be analyzed ...
12
13
We are running our first-ever datathon! Everyone has collected lots of data ... ready to be analyzed ...


2024
2024 [in photos]
Recent features (available online)

New methods in paleobiology | Women of Impact
Biomolecule & biosignature fossilization
Astrobiology
Paleobiology
Photo Gallery

"...to boldly go where no man has gone before": we find our samples for laboratory analyses in zoological, botanical, paleontological, and geological museum collections or collect them in the field! Here are a couple of photos from previous field expeditions and other adventurous trips!